PubMed API in Matlab#
by Anastasia Ramig
These recipe examples were tested on November 23, 2022 in MATLAB R2022b.
NCBI Entrez Programming Utilities Documentation: https://www.ncbi.nlm.nih.gov/books/NBK25501/
Please see NCBI’s Data Usage Policies and Disclaimers: https://www.ncbi.nlm.nih.gov/home/about/policies/
1. Basic PubMed API call#
For calling individual articles and publications, we will need to use this API URL:
%% set up the API parameters
summary = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esummary.fcgi?db=pubmed&";
The article we are requesting has PubMed ID: 27933103.
retmode in the web API URL specifies the file format. In this example, we will use JSON.
%% pull specific article data using its ID
url = summary + "id=27933103&retmode=json";
data_call = webread(url);
%% index to where the authors are stored
index = data_call.result.x27933103.authors;
disp(index)
Output:
8×1 struct array with fields:
name
authtype
clusterid
%% index to pull out the list of author names
names = {index(:).name};
disp(names)
Output:
Columns 1 through 5
{'Scalfani VF'} {'Williams AJ'} {'Tkachenko V'} {'Karapetyan K'} {'Pshenichnov A'}
Columns 6 through 8
{'Hanson RM'} {'Liddie JM'} {'Bara JE'}
2. Request Data using a Loop#
First, create a list of PubMed IDs:
%% create a list of PubMed IDs
idList = [34813985, 34813932, 34813684, 34813661, 34813372, 34813140, 34813072];
We will use map containers to store IDs and associated data. MATLAB map containers work by storing a value and associating it with a specific key. We can establish a map container using:
%% create an empty map container
map = containers.Map;
map
Output:
map =
Map with properties:
Count: 0
KeyType: char
ValueType: any
%% create two structures and add each id and its corresponding search
multiPapersKeys = {ones(length(idList), 1)};
multiPapersValues = {ones(length(idList), 1)};
for i=1:length(idList)
url = summary + "id=" + string(idList(i)) + "&retmode=json";
multiPapersKeys{i} = idList(i);
multiPapersValues{i} = webread(url);
pause(1)
end
%% create a container of the search results and index to a specific article
multiPapers = containers.Map(multiPapersKeys, multiPapersValues);
multiPapers(34813985)
Output:
ans = struct with fields:
header: [1×1 struct]
result: [1×1 struct]
%% create a new set of ids that are formatted with "x" for indexing
xiSet = {ones(length(idList),1)};
for i=1:length(idList)
xiSet{i} = "x" + idList(i);
end
%% get the title for each journal
for i=1:length(idList)
id = idList(i);
displayResult = multiPapers(id).result.(xiSet{i}).source
end
Output:
displayResult = 'Cell Calcium'
displayResult = 'Methods'
displayResult = 'FEBS J'
displayResult = 'Dev Growth Differ'
displayResult = 'CRISPR J'
displayResult = 'Chembiochem'
displayResult = 'Methods Mol Biol'
3. PubMed API Calls with Requests and Parameters#
For searching for articles using search term(s), we will need to use this API URL:
%% set the search url for the API
search = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=pubmed&";
When searching through articles, we are given a few ways of filtering the data. A list of all the available parameters for these requests can be found in the official NCBI documentation:
https://www.ncbi.nlm.nih.gov/books/NBK25499/
We can specify the database by putting db=<database> into the URL. We will be using the PubMed database. We can also use term to search data by adding term=<searchQuery>. Just be sure to replace spaces with a + instead. We can, for example, use a query to search PubMed, such as “neuroscience intervention learning”:
%% search the API
url = search + "term=neuroscience+intervention+learning&retmode=json";
data = webread(url);
The number of returned IDs can be adjusted with the retmax parameter:
%% limit the search to 25 articles and pull the list of ids
url = search + "term=neuroscience+intervention+learning&retmax=25&retmode=json";
data = webread(url);
disp(data.esearchresult.idlist)
Output:
{'36416175'}
{'36415971'}
{'36414247'}
{'36414012'}
{'36411719'}
{'36411683'}
{'36411673'}
{'36409100'}
{'36409046'}
{'36408530'}
{'36408399'}
{'36408106'}
{'36408061'}
{'36405490'}
{'36405191'}
{'36405080'}
{'36404677'}
{'36404570'}
{'36402843'}
{'36402815'}
{'36402739'}
{'36402496'}
{'36401545'}
{'36399451'}
{'36398842'}
length(data.esearchresult.idlist)
Output:
ans = 25
We can also use the query to search for an author. Add [au] after the name to specify it is an author.
%% search articles by author name
url = search+"term=Darwin[au]&retmode=json";
data = webread(url);
data.esearchresult.count
Output:
ans = '603'
We can also sort results using usehistory=y. This allows us to store the data for it to be sorted in the same API call. The addition of sort=pub+date will sort IDs by the publishing date.
%% perform a search that is sorted by publication date
url = search+"term=Coral+Reefs&retmode=json&usehistory=y&sort=pub+date";
data = webread(url);
disp(data.esearchresult.idlist)
Output:
{'35341677'}
{'36252668'}
{'36183766'}
{'36181819'}
{'36055494'}
{'35995149'}
{'36409983'}
{'36265239'}
{'36179999'}
{'36172974'}
{'36168958'}
{'36152066'}
{'36150619'}
{'36129389'}
{'36106689'}
{'36064010'}
{'36054745'}
{'35998799'}
{'35980514'}
{'35718641'}
%% compare to unsorted
url = search+"term=Coral+Reefs&retmode=json";
data = webread(url);
disp(data.esearchresult.idlist)
Output:
{'36416762'}
{'36415309'}
{'36413112'}
{'36409983'}
{'36406938'}
{'36405638'}
{'36401956'}
{'36401815'}
{'36399057'}
{'36395713'}
{'36395226'}
{'36389413'}
{'36385270'}
{'36383546'}
{'36382375'}
{'36379970'}
{'36379169'}
{'36372339'}
{'36371949'}
{'36371558'}
We can also search based on publication type by adding AND into the search in the term: term=<searchQuery>+AND+filter[filterType].
[pt] specifies that the filter type is the publication type. More filters can be found at: https://pubmed.ncbi.nlm.nih.gov/help/.
%% search based on publication type
url = search+"term=stem+cells+AND+clinical+trial[pt]&retmode=json";
data = webread(url)
Output:
data = struct with fields:
header: [1×1 struct]
esearchresult: [1×1 struct]
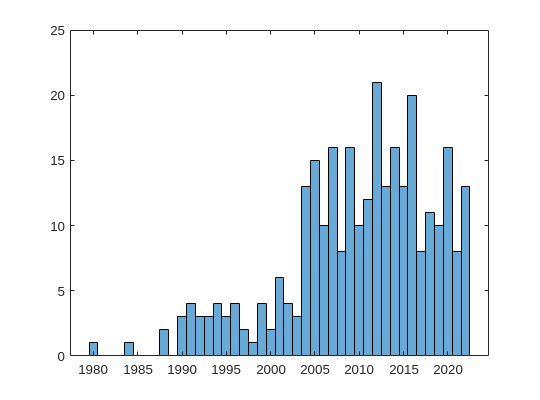
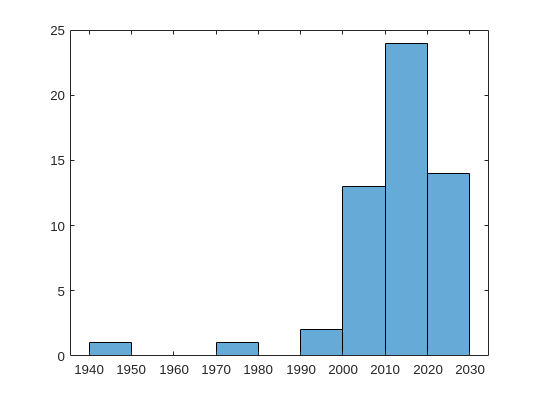
4. PubMed API Metadata Visualization#
Frequency of Topic sortpubdate field#
Extracting the sortpubdate field for a “hydrogel drug” search results, limited to publication type clinical trials:
%% perform a search using the term "hydrogel drug" and print the list of ids
url = search+"term=hydrogel+drug+AND+clinical+trial[pt]&sort=pub+date&retmax=500&retmode=json";
data = webread(url);
ids = data.esearchresult.idlist;
length(ids)
Output:
ans = 299
%% create a list of publication dates
pubDates = {ones(length(ids), 1)};
for i=1:length(ids)
url = summary+"id="+string(ids{i})+"&retmode=json";
request = webread(url);
pause(1)
idNew = "x" + ids{i};
pubDates{i} = request.result.(idNew).sortpubdate;
end
pubDates{1:10}
Output:
ans = '2022/12/01 00:00'
ans = '2022/10/19 00:00'
ans = '2022/10/01 00:00'
ans = '2022/10/01 00:00'
ans = '2022/08/01 00:00'
ans = '2022/06/01 00:00'
ans = '2022/05/01 00:00'
ans = '2022/04/01 00:00'
ans = '2022/03/01 00:00'
ans = '2022/01/21 00:00'
length(pubDates)
Output:
ans = 299
%% pull the year from each publication date
datesList = {ones(length(pubDates), 1)};
for i = 1:length(pubDates)
datesList{i} = str2double(pubDates{i}(1:4));
end
disp(datesList(1:20)) %% show first 20
Output:
Columns 1 through 8
{[2022]} {[2022]} {[2022]} {[2022]} {[2022]} {[2022]} {[2022]} {[2022]}
Columns 9 through 16
{[2022]} {[2022]} {[2022]} {[2022]} {[2022]} {[2021]} {[2021]} {[2021]}
Columns 17 through 20
{[2021]} {[2021]} {[2021]} {[2021]}
%% plot a histogram of the publications according to the decade in which they were published
x = cell2mat(datesList);
f = figure;
f.Position = [100 100 540 400];
f(1);
edges = [1980 1985 1990 1995 2000 2005 2010 2015 2020];
histogram(x)
Output: