PubChem API in R#
by Vishank Patel and Adam M. Nguyen
Documentation:
Pubchem API Documentation: https://pubchemdocs.ncbi.nlm.nih.gov/programmatic-access
These recipe examples were tested on March 24, 2023.
See the bottom of the document for information on R and package versions.
Attribution: This tutorial was adapted from supporting information in:
Scalfani, V. F.; Ralph, S. C. Alshaikh, A. A.; Bara, J. E. Programmatic Compilation of Chemical Data and Literature From PubChem Using Matlab. Chemical Engineering Education, 2020, 54, 230. https://doi.org/10.18260/2-1-370.660-115508 and vfscalfani/MATLAB-cheminformatics)
Setup#
Importing the necessary libraries and setting up the base api:
library(tidyverse) #essential packages
library(dplyr) #tibbles (R data_frames)
library(purrr) #character manipulation
library(httr) #GET() API requests
library(jsonlite) #converting to JSON
library(knitr) #including graphics
library(imager) #including images
library(magick) #Image manipulation
# Create base URL for PubChem API
api <- 'https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/'
1. PubChem Similarity#
Search for chemical structures in PubChem via a Fingerprint Tanimoto Similarity Search.
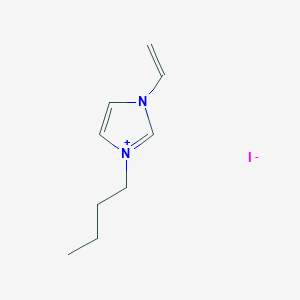
Get compound image#
compoundID <- "2734162"
CID_URL <- paste0(api,"cid/",compoundID,"/PNG") #paste0 concatenates strings
include_graphics(CID_URL)
Replace the above CID value (CID_SS_query) with a different CID to customize.
Retrieve InChI and SMILES#
Retrieve InChI
inchi_url <- paste0(api,"cid/",compoundID,"/property/inchi/TXT")
raw_inchi <- rawToChar(GET(inchi_url)$content); #"$content" filters the http response from the output and only returns the required output data
inchi <- raw_inchi %>% gsub("\n","",.); #"." refers to raw_inchi in gsub
inchi
## [1] "InChI=1S/C8H15N2/c1-3-4-5-10-7-6-9(2)8-10/h6-8H,3-5H2,1-2H3/q+1"
Retrieve Isomeric SMILES
IS_url <- paste0(api,"cid/",compoundID,"/property/IsomericSMILES/TXT");
raw_IS <- rawToChar(GET(IS_url)$content);
IS <- raw_IS %>% gsub("\n","",.);
IS
## [1] "CCCCN1C=C[N+](=C1)C"
Perform a Similarity Search#
Search for chemical structures by Similarity Search (SS), (2D Tanimoto threshold 95% to 1-Butyl-3-methyl-imidazolium; CID = 2734162)
api <- "https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/";
SS_url <- paste0(api,'fastsimilarity_2d/cid/',compoundID, '/cids/JSON?Threshold=95');
raw_output <- GET(SS_url)$content; #getting the unicode output
raw_result <- rawToChar(raw_output); #converts the output from unicode to text
CIDs1_ls <- fromJSON(raw_result); #creates a list of lists from the JSON data
head(CIDs1_ls)
## $IdentifierList
## $IdentifierList$CID
## [1] 2734161 61347 529334 304622 118785 12971008 11448496
## [8] 11424151 11171745 11160028 2734236 11245926 2734162 53384410
## [15] 11788435 5245884 2734168 139254006 91983981 87560886 87559770
## [22] 11448364 10537570 10154187 156579812 141109628 132087981 129831898
## [29] 118120095 91210418 90744066 87754289 87106874 87105369 37818425
## [36] 25053226 24766551 17870330 16720567 15557008 15255204 12392681
## [43] 12392676 11277167 11031767 10608883 10513048 10313448 10313447
## [50] 4183883 166063320 162198727 162171380 161819746 161124984 161112590
## [57] 160933286 160731515 160674142 160464403 160266994 159865990 159563905
## [64] 159509762 159490164 159312353 158808695 158224178 158212500 157839501
## [71] 157806980 157754871 157540783 157223666 157050789 156847779 156847755
## [78] 156447427 155713168 154005528 153940412 153897980 153615844 145473275
## [85] 145394036 144530620 144439506 144220344 143565409 143530527 142964175
## [92] 142585407 142152870 142141931 142071419 142062823 141885951 141442188
## [99] 141391260 141319938 141319527 141292180 141130209 140729180 140162362
## [106] 140162361 139215657 134991434 134345956 131201342 129864984 126970742
## [113] 124094863 124013672 124013100 123725669 122625623 121299516 118952202
## [120] 118524186 118524174 118131665 118057427 117890836 117703152 117684660
## [127] 102147231 101839829 101369383 91367525 90912888 89713026 89678233
## [134] 89432682 88864524 88236103 87942618 87813936 87806569 87790333
## [141] 87789992 87789923 87789740 87754264 87690425 87688227 87669070
## [148] 87572549 87572548 87572214 87572213 87560545 87560544 87509019
## [155] 87397668 87388314 87356647 87325711 87308565 87222859 87181405
## [162] 87181202 87181191 87181050 87173651 87125511 87125508 87121545
## [169] 87121544 87121543 87121443 87121324 87121318 87121317 87121316
## [176] 87121297 87121296 87121295 87099925 87096071 87092336 87059628
## [183] 86222771 69317070 68379078 67674484 66751376 60860613 60103428
## [190] 59872702 59798517 59672181 59653228 59653227 59161437 58850418
## [197] 57320166 54694426 54390693 54061781 53830935 53712415 25136370
## [204] 25030218 22382810 22055176 20646157 20646140 20612472 20532437
## [211] 20148470 19964884 19964723 19964469 17870312 16726386 14647320
## [218] 13723913 13034366 12392683 12392680 12392679 12392671 12392670
## [225] 12224822 11788445 11564856 11031768 10057718 11108531 11356498
## [232] 11390126 11390127 11404132 13034385 17885728 44178740 44178741
## [239] 44178743 44178745 45112083 54589973 54589974 54589975 54589976
## [246] 54589977 54589978 54589979 54589980 54589981 54589982 56973040
## [253] 57483533 60103429 66560137 66560138 68569453 87121298 87121319
## [260] 87121326 87121546 87125509 87338472 87457441 87645661 101456096
## [267] 101476671 101540709 101548772 102490458 102490464 117064740 123927694
## [274] 124705578 129761517 130557690 131038087 132565110 132604341 141130203
## [281] 142734045 149791673 155683104 156847757 165390285
Working with list of lists is hard, so we convert the same into a tibble. The code is adopted from: https://www.r-bloggers.com/2015/11/accessing-apis-from-r-and-a-little-r-programming/
CIDs1_df <- do.call( # do.call feeds in all the arguments (dataframes to be binded together) to the rbind command
what = "rbind", # rbind binds all the dataframes together
args = lapply( # lapply applies its second argument (as.data.frame) to every element of the list in the first argument (result1_ls)
CIDs1_ls, as.data.frame)) # converts the list of lists to a dataframe
CIDs1_df <- tibble(CIDs1_df) # converting the dataset into a tibble (from dplyr)
CIDs1_df # displaying the first few elements of the data
## # A tibble: 285 × 1
## CID
## <int>
## 1 2734161
## 2 61347
## 3 529334
## 4 304622
## 5 118785
## 6 12971008
## 7 11448496
## 8 11424151
## 9 11171745
## 10 11160028
## # … with 275 more rows
In the above SS_url value, you can adjust to the desired Tanimoto threshold (i.e., 97, 90, etc.)
Retrieve Identifier and Property Data#
Create an identifier/property dataset from Similarity Search results.
Retrieve the following data from CID hit results: InChI, Isomeric SMILES, MW, Heavy Atom Count, Rotable Bond Count, and Charge
short_CIDs <- CIDs1_df$CID[1:25] #taking the first 25 CIDs from the similarity search results
#initializing the tibble
similarity_results_tibble <- tibble();
similarity_results_tibble <- add_column(similarity_results_tibble,
Compound_ID = "",
InChi = "",
IsoSMI = "",
MW = "",
Heavy_Atom_Count = "",
Rotatable_Bond_Count = "",
Charge = ""
);
for (CID in short_CIDs) {
#define the api calls:
api = 'https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/';
CID_InChI_url = paste0(api,'cid/',toString(CID),'/property/InChI/TXT');
CID_IsoSMI_url = paste0(api,'cid/',toString(CID),'/property/IsomericSMILES/TXT');
CID_MW_url = paste0(api,'cid/',toString(CID),'/property/MolecularWeight/TXT');
CID_HeavyAtomCount_url = paste0(api,'cid/',toString(CID),'/property/HeavyAtomCount/TXT');
CID_RotatableBondCount_url = paste0(api,'cid/',toString(CID),'/property/RotatableBondCount/TXT');
CID_Charge_url = paste0(api,'cid/',toString(CID),'/property/Charge/TXT');
#downloading the data
inchi_temp <- rawToChar(GET(CID_InChI_url)$content) %>% gsub("\n","",.);
Sys.sleep(1) # adding a delay for the PubChem server
isoSMI_temp <- rawToChar(GET(CID_IsoSMI_url)$content) %>% gsub("\n","",.);
Sys.sleep(1)
mw_temp <- rawToChar(GET(CID_MW_url)$content) %>% gsub("\n","",.);
Sys.sleep(1)
heavy_atom_count_temp <- rawToChar(GET(CID_HeavyAtomCount_url)$content) %>% gsub("\n","",.);
Sys.sleep(1)
rotatable_bond_count_temp <- rawToChar(GET(CID_RotatableBondCount_url)$content) %>% gsub("\n","",.);
Sys.sleep(1)
charge_temp <- rawToChar(GET(CID_Charge_url)$content) %>% gsub("\n","",.);
Sys.sleep(1)
#Appending the data in a tibble
similarity_results_tibble <- similarity_results_tibble %>%
add_row(
Compound_ID = toString(CID),
InChi = inchi_temp,
IsoSMI = isoSMI_temp,
MW = mw_temp,
Heavy_Atom_Count = heavy_atom_count_temp,
Rotatable_Bond_Count = rotatable_bond_count_temp,
Charge = charge_temp
)
}
similarity_results_tibble
## # A tibble: 25 × 7
## Compound_ID InChi IsoSMI MW Heavy…¹ Rotat…² Charge
## <chr> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 2734161 InChI=1S/C8H15N2.ClH/c1-3-4-… CCCCN… 174.… 11 3 0
## 2 61347 InChI=1S/C7H12N2/c1-2-3-5-9-… CCCCN… 124.… 9 3 0
## 3 529334 InChI=1S/C8H14N2/c1-2-3-4-6-… CCCCC… 138.… 10 4 0
## 4 304622 InChI=1S/C8H14N2/c1-3-4-6-10… CCCCN… 138.… 10 3 0
## 5 118785 InChI=1S/C6H10N2/c1-2-4-8-5-… CCCN1… 110.… 8 2 0
## 6 12971008 InChI=1S/C7H13N2.HI/c1-3-4-9… CCCN1… 252.… 10 2 0
## 7 11448496 InChI=1S/C8H15N2.HI/c1-3-4-5… CCCCN… 266.… 11 3 0
## 8 11424151 InChI=1S/C8H15N2.CHNS/c1-3-4… CCCCN… 197.… 13 3 0
## 9 11171745 InChI=1S/C8H15N2.C2N3/c1-3-4… CCCCN… 205.… 15 3 0
## 10 11160028 InChI=1S/C7H13N2.BrH/c1-3-4-… CCCN1… 205.… 10 2 0
## # … with 15 more rows, and abbreviated variable names ¹Heavy_Atom_Count,
## # ²Rotatable_Bond_Count
We will now export the generated dataframe as a tab separated text file. The file will be saved in the present working directory.
write.table(similarity_results_tibble, file = "Data/R_Similarityq_results.txt", sep = "\t", row.names = TRUE, col.names = NA);
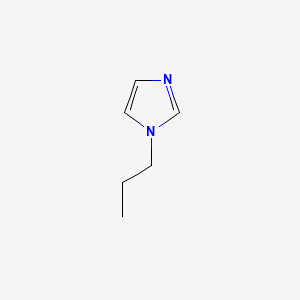
Retrieve Images of CID Compounds from Similarity Search#
Create the results png:
content <- list()
for(CID in short_CIDs[1:5]){
#define the url
CID_URL <- paste0(api,"cid/",toString(CID),"/PNG");
content[[length(content) + 1]] <- GET(CID_URL)$content
Sys.sleep(1)
}
image_read(content[[1]])
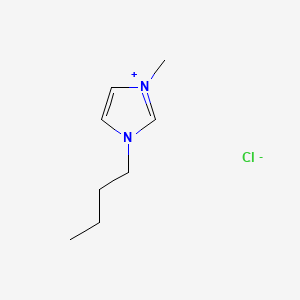
image_read(content[[2]])
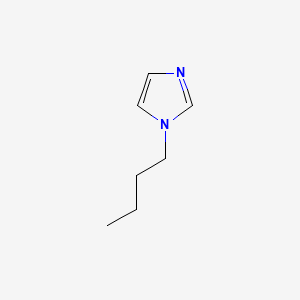
image_read(content[[3]])
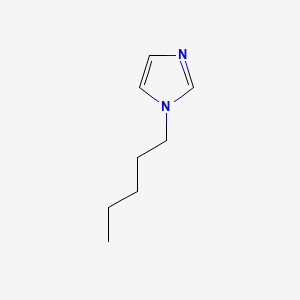
image_read(content[[4]])
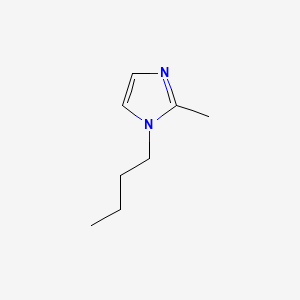
image_read(content[[5]])
2. PubChem SMARTS Search#
Search for chemical structures in PubChem via a SMARTS substructure query and compile results
#defining the PubChem base API
api <- "https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/";
Define SMARTS Queries#
# view pattern syntax at: https://smartsview.zbh.uni-hamburg.de/
# these are vinyl imidazolium substructure searches
SmartsQ <- c("[CR0H2][n+]1[cH1][cH1]n([CR0H1]=[CR0H2])[cH1]1","[CR0H2][n+]1[cH1][cH1]n([CR0H2][CR0H1]=[CR0H2])[cH1]1","[CR0H2][n+]1[cH1][cH1]n([CR0H2][CR0H2][CR0H1]=[CR0H2])[cH1]1");
Add your own SMARTS queries to customize. You can add as many as desired within a cell array.
Perform a SMARTS Query Search#
#generate URLs for SMARTS query searches
SmartsQ_url= c();
for (Smarts_query in SmartsQ) {
SmartsQ_url <- append(SmartsQ_url,
values = paste0(api,"fastsubstructure/smarts/",Smarts_query,"/cids/JSON"));
}
#perform substructure searches for each query link in SMARTSq_url
hit_CIDs_results_seperate =c();
for (url in SmartsQ_url) {
hit_CIDs_ls_temp <- fromJSON(rawToChar(GET(url)$content))
hit_CIDs_df_temp <- tibble(do.call(what = "rbind",
args = lapply(hit_CIDs_ls_temp, as.data.frame)))
hit_CIDs_results_seperate <- append(hit_CIDs_results_seperate, hit_CIDs_df_temp)
}
#create the final result by appending each of the returned CID list outputs
hit_CIDs_results <- c();
hit_CIDs_results <- append(hit_CIDs_results,
values = c(hit_CIDs_results_seperate[1]$CID,
hit_CIDs_results_seperate[2]$CID,
hit_CIDs_results_seperate[3]$CID))
length(hit_CIDs_results)
## [1] 847
We will shorten the list to the first 25 results for time considerations. This limit can be increased.
hit_CIDs_results <- hit_CIDs_results[1:25];
hit_CIDs_results
## [1] 121235111 132274871 86657882 46178576 139254006 129853306 129853221
## [8] 129850195 87560886 87559770 87327009 59435292 24766550 162277521
## [15] 162204473 161909937 161556919 161457090 161355970 161152181 161033043
## [22] 160979882 160848969 160787397 160707335
Retrieve Identifier and Property Data#
Create an identifier/property dataset from the SMARTS substructure search results
Retrieve the following data for each CID: InChI, Canonical SMILES, MW, IUPAC Name, Heavy Atom Count, Covalent Unit Count, Charge
smarts_results_tibble <- tibble();
smarts_results_tibble <- add_column(smarts_results_tibble,
Compound_ID = "",
InChi = "",
CanSMI = "",
MW = "",
IUPACName = "",
Heavy_Atom_Count = "",
Covalent_Unit_Count = "",
Charge = ""
);
for (CID in hit_CIDs_results) {
#define the api calls:
api = 'https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/';
CID_InChI_url = paste0(api,'cid/',toString(CID),'/property/InChI/TXT');
CID_CanSMI_url = paste0(api,'cid/',toString(CID),'/property/CanonicalSMILES/TXT');
CID_MW_url = paste0(api,'cid/',toString(CID),'/property/MolecularWeight/TXT');
CID_IUPACName_url = paste0(api,'cid/',toString(CID),'/property/IUPACName/TXT');
CID_HeavyAtomCount_url = paste0(api,'cid/',toString(CID),'/property/HeavyAtomCount/TXT');
CID_CovalentUnitCount_url = paste0(api,'cid/',toString(CID),'/property/CovalentUnitCount/TXT');
CID_Charge_url = paste0(api,'cid/',toString(CID),'/property/Charge/TXT');
#downloading the data
inchi_temp <- rawToChar(GET(CID_InChI_url)$content) %>% gsub("\n","",.);
Sys.sleep(1) # adding a delay for the PubChem server
canSMI_temp <- rawToChar(GET(CID_CanSMI_url)$content) %>% gsub("\n","",.);
Sys.sleep(1)
mw_temp <- rawToChar(GET(CID_MW_url)$content) %>% gsub("\n","",.);
Sys.sleep(1)
iupac_name_temp <- rawToChar(GET(CID_IUPACName_url)$content) %>% gsub("\n","",.);
Sys.sleep(1)
heavy_atom_count_temp <- rawToChar(GET(CID_HeavyAtomCount_url)$content) %>% gsub("\n","",.);
Sys.sleep(1)
covalent_unit_count_temp <- rawToChar(GET(CID_CovalentUnitCount_url)$content) %>% gsub("\n","",.);
Sys.sleep(1)
charge_temp <- rawToChar(GET(CID_Charge_url)$content) %>% gsub("\n","",.);
Sys.sleep(1)
#Appending the data in a tibble
smarts_results_tibble <- smarts_results_tibble %>%
add_row(
Compound_ID = toString(CID),
InChi = inchi_temp,
CanSMI = canSMI_temp,
MW = mw_temp,
IUPACName = iupac_name_temp,
Heavy_Atom_Count = heavy_atom_count_temp,
Covalent_Unit_Count = covalent_unit_count_temp,
Charge = charge_temp
)
}
smarts_results_tibble
## # A tibble: 25 × 8
## Compound_ID InChi CanSMI MW IUPAC…¹ Heavy…² Coval…³ Charge
## <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 121235111 InChI=1S/C7H11N2.C2F… CC[N+… 403.3 bis(tr… 24 2 0
## 2 132274871 InChI=1S/C14H20N4.2C… C=CN1… 804.6 bis(tr… 48 3 0
## 3 86657882 InChI=1S/C13H23N2.Br… CCCCC… 287.… 1-ethe… 16 2 0
## 4 46178576 InChI=1S/C21H39N2.Br… CCCCC… 399.5 1-ethe… 24 2 0
## 5 139254006 InChI=1S/C9H15N2.HI/… CCCC[… 278.… 1-buty… 12 2 0
## 6 129853306 InChI=1S/C13H23N2.C2… CCCCC… 487.5 bis(tr… 30 2 0
## 7 129853221 InChI=1S/C11H19N2.C2… CCCCC… 459.4 bis(tr… 28 2 0
## 8 129850195 InChI=1S/C21H39N2.C2… CCCCC… 599.7 bis(tr… 38 2 0
## 9 87560886 InChI=1S/C9H15N2.BrH… CCCC[… 231.… 1-buty… 12 2 0
## 10 87559770 InChI=1S/C9H15N2.ClH… CCCC[… 186.… 1-buty… 12 2 0
## # … with 15 more rows, and abbreviated variable names ¹IUPACName,
## # ²Heavy_Atom_Count, ³Covalent_Unit_Count
Rearrange the result’s columns:
smarts_results_tibble2 <- smarts_results_tibble[c("CanSMI","IUPACName","Compound_ID","InChi","MW","Heavy_Atom_Count","Covalent_Unit_Count","Charge")]
Exporting the data as a tabbed text file
write.table(smarts_results_tibble2, file = "Data/R_Smartsq_results.txt", sep = "\t", row.names = TRUE, col.names = NA);
Retrieve Images of CID Compounds from SMARTS query match#
Create the results png:
content2 <- list()
for(CID in hit_CIDs_results[1:5]){
#define the url
hit_CID_URL <- paste0(api,"cid/",toString(CID),"/PNG");
content2[[length(content2) + 1]] <- GET(hit_CID_URL)$content
Sys.sleep(1)
}
image_read(content2[[1]])
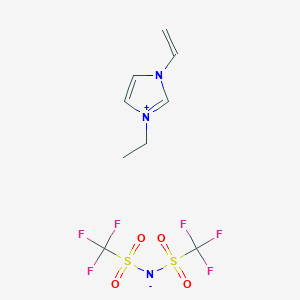
image_read(content2[[2]])
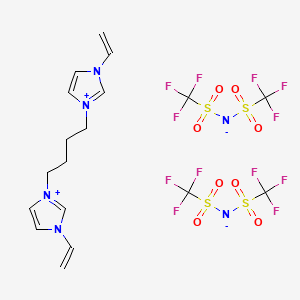
image_read(content2[[3]])
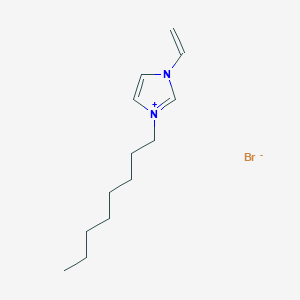
image_read(content2[[4]])
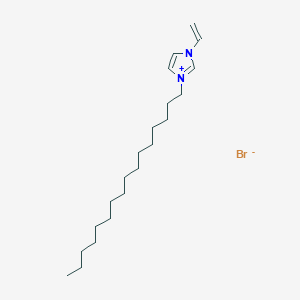
image_read(content2[[5]])
R Session Info#
sessionInfo()
## R version 4.2.1 (2022-06-23 ucrt)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 19042)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=English_United States.utf8
## [2] LC_CTYPE=English_United States.utf8
## [3] LC_MONETARY=English_United States.utf8
## [4] LC_NUMERIC=C
## [5] LC_TIME=English_United States.utf8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] magick_2.7.4 imager_0.42.18 magrittr_2.0.3 knitr_1.42
## [5] jsonlite_1.8.4 httr_1.4.5 lubridate_1.9.2 forcats_1.0.0
## [9] stringr_1.5.0 dplyr_1.1.0 purrr_1.0.1 readr_2.1.4
## [13] tidyr_1.3.0 tibble_3.1.8 ggplot2_3.4.1 tidyverse_2.0.0
##
## loaded via a namespace (and not attached):
## [1] tidyselect_1.2.0 xfun_0.37 bslib_0.4.2 colorspace_2.1-0
## [5] vctrs_0.5.2 generics_0.1.3 htmltools_0.5.4 yaml_2.3.7
## [9] utf8_1.2.3 rlang_1.0.6 jquerylib_0.1.4 pillar_1.8.1
## [13] glue_1.6.2 withr_2.5.0 jpeg_0.1-10 lifecycle_1.0.3
## [17] munsell_0.5.0 gtable_0.3.1 evaluate_0.20 tzdb_0.3.0
## [21] fastmap_1.1.0 curl_5.0.0 fansi_1.0.4 highr_0.10
## [25] Rcpp_1.0.10 scales_1.2.1 cachem_1.0.7 hms_1.1.2
## [29] bmp_0.3 png_0.1-8 digest_0.6.31 stringi_1.7.12
## [33] tiff_0.1-11 grid_4.2.1 cli_3.6.0 tools_4.2.1
## [37] sass_0.4.5 readbitmap_0.1.5 pkgconfig_2.0.3 ellipsis_0.3.2
## [41] timechange_0.2.0 rmarkdown_2.20 rstudioapi_0.14 R6_2.5.1
## [45] igraph_1.4.1 compiler_4.2.1
