PubChem API in Unix Shell#
by Avery Fernandez and Vincent F. Scalfani
These recipe examples were tested on August 4, 2022 using GNOME Terminal (with Bash 4.4.20) in Ubuntu 18.04.
PubChem API Documentation: https://pubchemdocs.ncbi.nlm.nih.gov/programmatic-access
Attribution: This tutorial was adapted from supporting information in:
Scalfani, V. F.; Ralph, S. C. Alshaikh, A. A.; Bara, J. E. Programmatic Compilation of Chemical Data and Literature From PubChem Using Matlab. Chemical Engineering Education, 2020, 54, 230. https://doi.org/10.18260/2-1-370.660-115508 and vfscalfani/MATLAB-cheminformatics)
Note
This tutorial uses curl and jq for interacting with the PubChem API. You may also be interested in using the NCBI EDirect command line program. We have several tutorials for EDirect in our EDirectChemInfo repository.
Setup#
Program requirements#
In order to run this code, you will need to first install curl, and jq. curl is used to request the data from the API, and jq is used to parse the JSON data. In addition, if you want to be able to print the molecules as ASCII characters in your terminal, you will need to install RDKit and download the print_mols Python script.
Define base URL#
Define the PubChem PUG-REST API base URL:
api="https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/"
1. PubChem Similarity#
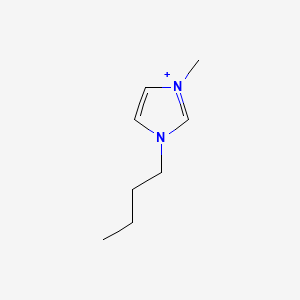
Get Compound Image#
We can search for a compound and display an image, for example: 1-Butyl-3-methyl-imidazolium; CID = 2734162
compoundID="2734162"
curl -s "$api"$"cid/""$compoundID"$"/PNG" -o CID_2734162.png
Note
The silent option (-s) for curl was used to hide the progress outputs.
If you want to open the PNG file in an image viewer program from your terminal, try xdg-open:
xdg-open CID_2734162.png
Output:
Retrieve InChI and SMILES#
request=$(curl -s "$api""cid/""$compoundID""/property/inchi,IsomericSMILES/JSON")
echo "$request" | jq '.'
Output:
{
"PropertyTable": {
"Properties": [
{
"CID": 2734162,
"IsomericSMILES": "CCCCN1C=C[N+](=C1)C",
"InChI": "InChI=1S/C8H15N2/c1-3-4-5-10-7-6-9(2)8-10/h6-8H,3-5H2,1-2H3/q+1"
}
]
}
}
Now, extract out the InChI:
echo "$request" | jq '.["PropertyTable"]["Properties"][0]["InChI"]'
Output:
"InChI=1S/C8H15N2/c1-3-4-5-10-7-6-9(2)8-10/h6-8H,3-5H2,1-2H3/q+1"
And the IsomericSMILES:
echo "$request" | jq '.["PropertyTable"]["Properties"][0]["IsomericSMILES"]'
Output:
"CCCCN1C=C[N+](=C1)C"
Display Molecule as ASCII Drawing#
We can use the extracted SMILES to generate an ASCII drawing within our terminal. First, we will extract the SMILES using jq, and then pipe the SMILES to a print_mols Python script, which uses the cheminformatics program RDKit to parse the SMILES, compute drawing coordinates, and then print the molecule as ASCII characters:
echo "$request" | jq '.["PropertyTable"]["Properties"][0]["IsomericSMILES"]' | tr -d '"' | python3 print_mols.py -
Output:
C
*
C *
* N
*
C C N * C
* * *
C * * C
C
Note
tr -d '"' removes the quotes around the extracted SMILES; python3 print_mols.py - prints the molecule.
Perform a Similarity Search#
We will use the PubChem API to perform a Fingerprint Tanimoto Similarity Search.
(2D Tanimoto threshold 95% to 1-Butyl-3-methyl-imidazolium; CID = 2734162)
request=$(curl -s "$api"$"fastsimilarity_2d/cid/""$compoundID"$"/cids/JSON?Threshold=95")
In the above request value, you can adjust to the desired Tanimoto threshold (e.g., 97, 90, etc.)
Let’s find the number of CID values returned and display the first 10 CIDs:
length=$(echo "$request" | jq '.["IdentifierList"]["CID"] | length')
echo "$length"
Output:
283
echo "$request" | jq ".IdentifierList.CID[0:10]"
Output:
[
2734161,
61347,
529334,
304622,
118785,
12971008,
11448496,
11424151,
11171745,
11160028
]
Note
Here is another approach to displaying the first 10 lines: echo "$request" | jq '.["IdentifierList"]["CID"]' | head -n10
Next, we will convert the CID identifier values into an array:
declare -a idList
for (( i = 0 ; i < length ; i++));
do
idList+=(" $(echo "$request" | jq ".IdentifierList.CID[$i]") ")
done
Display the first 10:
echo "${idList[@]:0:10}"
Output:
2734161 61347 529334 304622 118785 12971008 11448496 11424151 11171745 11160028
Retrieve Identifier and Property Data#
Get the following data for the retrieved CIDs (idList): InChI, Isomeric SMILES, MW, Heavy Atom Count, Rotable Bond Count, and Charge. As a test, we will only get data for the first 5 CIDs:
for id in "${idList[@]:0:5}"
do
compound=$(echo "$id" | sed 's/ //g')
request=$(curl -s "$api"$"cid/""$compound"$"/property/InChI,IsomericSMILES,MolecularWeight,HeavyAtomCount,RotatableBondCount,Charge/JSON")
echo "$request" | jq '.["PropertyTable"]["Properties"][0]'
sleep 1
done
Output:
{
"CID": 2734161,
"MolecularWeight": "174.67",
"IsomericSMILES": "CCCCN1C=C[N+](=C1)C.[Cl-]",
"InChI": "InChI=1S/C8H15N2.ClH/c1-3-4-5-10-7-6-9(2)8-10;/h6-8H,3-5H2,1-2H3;1H/q+1;/p-1",
"Charge": 0,
"RotatableBondCount": 3,
"HeavyAtomCount": 11
}
{
"CID": 61347,
"MolecularWeight": "124.18",
"IsomericSMILES": "CCCCN1C=CN=C1",
"InChI": "InChI=1S/C7H12N2/c1-2-3-5-9-6-4-8-7-9/h4,6-7H,2-3,5H2,1H3",
"Charge": 0,
"RotatableBondCount": 3,
"HeavyAtomCount": 9
}
{
"CID": 529334,
"MolecularWeight": "138.21",
"IsomericSMILES": "CCCCCN1C=CN=C1",
"InChI": "InChI=1S/C8H14N2/c1-2-3-4-6-10-7-5-9-8-10/h5,7-8H,2-4,6H2,1H3",
"Charge": 0,
"RotatableBondCount": 4,
"HeavyAtomCount": 10
}
{
"CID": 304622,
"MolecularWeight": "138.21",
"IsomericSMILES": "CCCCN1C=CN=C1C",
"InChI": "InChI=1S/C8H14N2/c1-3-4-6-10-7-5-9-8(10)2/h5,7H,3-4,6H2,1-2H3",
"Charge": 0,
"RotatableBondCount": 3,
"HeavyAtomCount": 10
}
{
"CID": 118785,
"MolecularWeight": "110.16",
"IsomericSMILES": "CCCN1C=CN=C1",
"InChI": "InChI=1S/C6H10N2/c1-2-4-8-5-3-7-6-8/h3,5-6H,2,4H2,1H3",
"Charge": 0,
"RotatableBondCount": 2,
"HeavyAtomCount": 8
}
Note
sed 's/ //g' removes the extra space before the CID values. tr -d ' ' should also work to remove the extra space.
We can modify the jq line to extract out specific data values such as the MolecularWeight:
for id in "${idList[@]:0:5}"
do
compound=$(echo "$id" | sed 's/ //g')
request=$(curl -s "$api"$"cid/""$compound"$"/property/InChI,IsomericSMILES,MolecularWeight,HeavyAtomCount,RotatableBondCount,Charge/JSON")
echo "$request" | jq '.["PropertyTable"]["Properties"][0]["MolecularWeight"]'
sleep 1
done
Output:
"174.67"
"124.18"
"138.21"
"138.21"
"110.16"
Retrieve Images of CID Compounds from Similarity Search#
We will get and save the PNG images for the first 5 compounds:
for id in "${idList[@]:0:5}"
do
compound=$(echo "$id" | sed 's/ //g')
request=$(curl -s "$api"$"cid/""$compound"$"/PNG" -o "$compound"$".png")
sleep 1
done
ls
Output:
118785.png 2734161.png 304622.png 529334.png 61347.png
Finally, we can also visualize the compounds as ASCII drawings using the print_mols Python script demonstrated above.
for id in "${idList[@]:0:5}"
do
compound=$(echo "$id" | sed 's/ //g')
request=$(curl -s "$api"$"cid/""$compound"$"/property/IsomericSMILES/JSON")
echo "$request" | jq '.["PropertyTable"]["Properties"][0]["IsomericSMILES"]' |
tr -d '"' |
python3 print_mols.py -
sleep 1
done
Output:
Cl
C * C
*
*
N * C
C C N *
* * * * *
C C C
C * N
*
*
C
C C N *
* * * * *
C C C
C * N
*
*
C
C C N *
* * * * * *
C C C C
C
*
C C
* * * * C
* *
C C N N
*
*
C
*
C
C * N
*
*
C
C N *
* * * *
C C C
2. PubChem SMARTS Search#
Search for chemical structures from a SMARTS substructure query.
Define SMARTS queries#
View pattern syntax at: https://smartsview.zbh.uni-hamburg.de/
Note: These are vinyl imidazolium substructure searches
declare -a smartsQ=("[CR0H2][n+]1[cH1][cH1]n([CR0H1]=[CR0H2])[cH1]1" "[CR0H2][n+]1[cH1][cH1]n([CR0H2][CR0H1]=[CR0H2])[cH1]1" "[CR0H2][n+]1[cH1][cH1]n([CR0H2][CR0H2][CR0H1]=[CR0H2])[cH1]1")
Add your own SMARTS queries to customize. You can add as many as desired within an array.
Perform a SMARTS query search#
We will combine all data into a single array:
declare -a combinedA
for smarts in "${smartsQ[@]}"
do
request=$(curl -s -g "$api"$"fastsubstructure/smarts/""$smarts"$"/cids/JSON")
sleep 1
length=$(echo "$request" | jq '.["IdentifierList"]["CID"] | length')
echo "$length"
for (( i = 0 ; i < length ; i++));
do
combinedA+=(" $(echo "$request" | jq ".IdentifierList.CID[$i]" ) ")
done
done
Output:
605
225
7
Note
The -g option with curl prevents curl from interpreting the [] characters in the SMARTS patterns.
Get length of array:
echo "${#combinedA[@]}"
Output:
837
Show the first 5 results:
echo "${combinedA[@]:0:5}"
Output:
121235111 86657882 46178576 24766550 139254006
Retrieve Identifier and Property Data#
We will retrieve some property data for the first 5 CIDs:
for id in "${combinedA[@]:0:5}"
do
compound=$(echo "$id" | tr -d ' ')
request=$(curl -s "$api"$"cid/""$compound"$"/property/InChI,CanonicalSMILES,MolecularWeight,IUPACName,HeavyAtomCount,CovalentUnitCount,Charge/JSON")
echo "$request" | jq '.["PropertyTable"]["Properties"][0]'
sleep 1
done
Output:
{
"CID": 121235111,
"MolecularWeight": "403.3",
"CanonicalSMILES": "CC[N+]1=CN(C=C1)C=C.C(F)(F)(F)S(=O)(=O)[N-]S(=O)(=O)C(F)(F)F",
"InChI": "InChI=1S/C7H11N2.C2F6NO4S2/c1-3-8-5-6-9(4-2)7-8;3-1(4,5)14(10,11)9-15(12,13)2(6,7)8/h3,5-7H,1,4H2,2H3;/q+1;-1",
"IUPACName": "bis(trifluoromethylsulfonyl)azanide;1-ethenyl-3-ethylimidazol-3-ium",
"Charge": 0,
"HeavyAtomCount": 24,
"CovalentUnitCount": 2
}
{
"CID": 86657882,
"MolecularWeight": "287.24",
"CanonicalSMILES": "CCCCCCCC[N+]1=CN(C=C1)C=C.[Br-]",
"InChI": "InChI=1S/C13H23N2.BrH/c1-3-5-6-7-8-9-10-15-12-11-14(4-2)13-15;/h4,11-13H,2-3,5-10H2,1H3;1H/q+1;/p-1",
"IUPACName": "1-ethenyl-3-octylimidazol-3-ium;bromide",
"Charge": 0,
"HeavyAtomCount": 16,
"CovalentUnitCount": 2
}
{
"CID": 46178576,
"MolecularWeight": "399.5",
"CanonicalSMILES": "CCCCCCCCCCCCCCCC[N+]1=CN(C=C1)C=C.[Br-]",
"InChI": "InChI=1S/C21H39N2.BrH/c1-3-5-6-7-8-9-10-11-12-13-14-15-16-17-18-23-20-19-22(4-2)21-23;/h4,19-21H,2-3,5-18H2,1H3;1H/q+1;/p-1",
"IUPACName": "1-ethenyl-3-hexadecylimidazol-3-ium;bromide",
"Charge": 0,
"HeavyAtomCount": 24,
"CovalentUnitCount": 2
}
{
"CID": 24766550,
"MolecularWeight": "431.4",
"CanonicalSMILES": "CCCC[N+]1=CN(C=C1)C=C.C(F)(F)(F)S(=O)(=O)[N-]S(=O)(=O)C(F)(F)F",
"InChI": "InChI=1S/C9H15N2.C2F6NO4S2/c1-3-5-6-11-8-7-10(4-2)9-11;3-1(4,5)14(10,11)9-15(12,13)2(6,7)8/h4,7-9H,2-3,5-6H2,1H3;/q+1;-1",
"IUPACName": "bis(trifluoromethylsulfonyl)azanide;1-butyl-3-ethenylimidazol-1-ium",
"Charge": 0,
"HeavyAtomCount": 26,
"CovalentUnitCount": 2
}
{
"CID": 139254006,
"MolecularWeight": "278.13",
"CanonicalSMILES": "CCCC[N+]1=CN(C=C1)C=C.[I-]",
"InChI": "InChI=1S/C9H15N2.HI/c1-3-5-6-11-8-7-10(4-2)9-11;/h4,7-9H,2-3,5-6H2,1H3;1H/q+1;/p-1",
"IUPACName": "1-butyl-3-ethenylimidazol-1-ium;iodide",
"Charge": 0,
"HeavyAtomCount": 12,
"CovalentUnitCount": 2
}
Get only the InChIs:
for id in "${combinedA[@]:0:5}"
do
compound=$(echo "$id" | tr -d ' ')
request=$(curl -s "$api"$"cid/""$compound"$"/property/InChI/JSON")
echo "$request" | jq '.["PropertyTable"]["Properties"][0]["InChI"]' | tr -d '"'
sleep 1
done
Output:
InChI=1S/C7H11N2.C2F6NO4S2/c1-3-8-5-6-9(4-2)7-8;3-1(4,5)14(10,11)9-15(12,13)2(6,7)8/h3,5-7H,1,4H2,2H3;/q+1;-1
InChI=1S/C13H23N2.BrH/c1-3-5-6-7-8-9-10-15-12-11-14(4-2)13-15;/h4,11-13H,2-3,5-10H2,1H3;1H/q+1;/p-1
InChI=1S/C21H39N2.BrH/c1-3-5-6-7-8-9-10-11-12-13-14-15-16-17-18-23-20-19-22(4-2)21-23;/h4,19-21H,2-3,5-18H2,1H3;1H/q+1;/p-1
InChI=1S/C9H15N2.C2F6NO4S2/c1-3-5-6-11-8-7-10(4-2)9-11;3-1(4,5)14(10,11)9-15(12,13)2(6,7)8/h4,7-9H,2-3,5-6H2,1H3;/q+1;-1
InChI=1S/C9H15N2.HI/c1-3-5-6-11-8-7-10(4-2)9-11;/h4,7-9H,2-3,5-6H2,1H3;1H/q+1;/p-1
